Group of Víctor Quesada and Pedro Robles
| Responsible | Víctor Quesada y Pedro Robles |
RESEARCH SUMMARY
We are performing in our laboratories a reverse genetic approach to identify and characterize Arabidopsis thaliana mutants affected in nuclear genes coding for proteins involved in the flow of genetic information in chloroplasts and mitochondria. For most of the studied genes, there have not been hitherto described any mutant phenotype resulting from impaired function. We are focused in two protein families: the mitochondrial transcription termination factors (mTERFs), and the chloroplast and mitochondrial ribosomal proteins (respectively cpRPs and mtRPs). Our main goal is to shed light on the understanding of the functions of mTERFs, cpRPs and MRPs in plant growth, development and adaptation to abiotic stress, especially to salinity.
RESEARCH LINES
Line 1: Genetic and molecular characterization of the Arabidopsis thaliana mitochondrial transcription termination factors (mTERFs)
The mTERF factors are coded by nuclear genes and in plants they carry out their functions in chloroplasts and mitochondria, where they are required for appropriate organellar gene expression. Land plant genomes include a much higher numbers of mTERF genes than animals. However, only a few of them have been molecularly characterized through the analysis of loss-of-function mutants. We are performing in our laboratories a phenotypic, genetic and molecular characterization of several Arabidopsis mterf mutants (e.g., mda1/mterf5, mterf6 and mterf9) which show different perturbations in growth and development, as well as an altered response to abiotic stress.
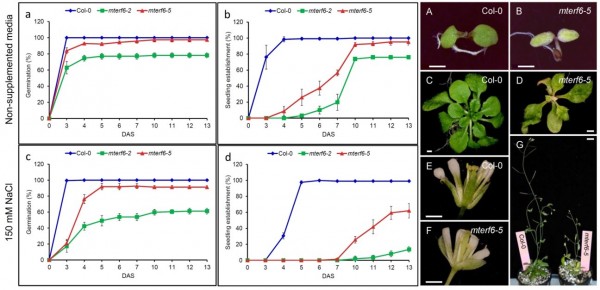
The mterf6 mutants (in red) are more sensitive to salinity stress (150 mM NaCl) than the wild-type Columbia-0 (Col-0; in blue) during germination and seedling establishment (a-d panels). Additionally, the mterf6-5 mutant shows growth, vegetative and reproductive developmental alterations (A-G panels on the right).
Line 2: Functional analysis of chloroplast and mitochondrial ribosomal proteins in Arabidopsis thaliana
Ribosomes, made of RNAs and proteins, are the molecular machinery that translate messenger RNAs in prokaryotes as well as and in the cytoplasm, chloroplasts and mitochondria of eukaryotic cells. With the aim of increasing the understanding of the functions of chloroplast and mitochondrial ribosomal proteins (respectively cpRPs and MRPs) in plants, we have initiated a reverse genetic approach to identify and characterize mutants affected in this kind of proteins in the plant model system Arabidopsis thaliana. We have identified mutant alleles carrying a T-DNA insertion in nuclear genes coding cpRPs or MRPs for which any loss-of-function viable alleles have been hitherto described. Currently we are characterizing these mutant at the phenotypic, genetic and molecular levels.

Rosette phenotypes of mutants affected in nuclear genes encoding chloroplast (upper panel) or mitochondrial (lower panel) ribosomal proteins. In the upper left corner it is shown a picture of the wild-type Columbia-0, the genetic background of all these mutants.
P. Robles, V. Quesada
Research Progress in the Molecular Functions of Plant mTERF Proteins
Cells 10, 205 (2021).
E. Núñez-Delegido, P. Robles, A. Ferrández-Ayela, V. Quesada
Plant Biology, 22, 459-471 (2020).
A. Lidón-Soto, E. Núñez-Delegido, I. Pastor-Martínez, P. Robles, V.Quesada
Arabidopsis plastid-RNA polymerase RPOTp is involved in abiotic stress tolerance
Plants 9, 834 (2020).
P. Robles P, V. Quesada
Transcriptional and post-transcriptional regulation of OGE and its roles in plant salt tolerance.
International Journal of Molecular Sciences, 20, 1056 (2019).
P. Robles, E. Núñez-Delegido, A. Ferrández-Ayela, R. Sarmiento-Mañús, J.L. Micol, V. Quesada
Arabidopsis mTERF6 is required for leaf patterning.
Plant Science 266, 117-129 (2018).
P. Robles, V. Quesada
International Journal of Molecular Sciences 22, 2104 (2021).
P. Robles, V. Quesada
Emerging Roles of Mitochondrial Ribosomal Proteins in Plant Development.
International Journal of Molecular Sciences, 18: 2595 (2017).
V. Quesada
The roles of mitochondrial transcription termination factors (MTERFs) in plants.
Physiologia Plantarum, 157, 389-399 (2016).
P. Robles, J.L. Micol, V. Quesada
Physiologia Plantarum 154, 297-313 (2015).
P. Robles, J.L. Micol, V. Quesada
PLoS ONE 8, e42924 (2012).
Caracterización genética y molecular de mutantes afectados en proteínas ribosómicas mitocondriales de Arabidopsis thaliana. Agencia de financiación: Universidad Miguel Hernández de Elche. Vigencia: desde: 2020 – 2021.
PI: Víctor Manuel Quesada Pérez.
Unidad de Microscopía de Fluorescencia de Hoja de Luz (IDIFEDER/2018/016). Instituto de Bioingeniería. Universidad Miguel Hernández. Agencia de financiación: Convocatoria de subvenciones para infraestructuras y equipamiento I+D+i de la Generalitat Valenciana. Vigencia: 2018 – 2020.
PI: José Manuel Pérez.
Regulación y nuevas funciones del gen ARGONAUTE1 de Arabidopsis”. Instituto de Bioingeniería. Universidad Miguel Hernández. Agencia de financiación: Ministerio de Economía y Competitividad. Vigencia: desde: 2015 – 2017.
PI: María Rosa Ponce.
Caracterización funcional de los genes mTERF MDA1 y MDA2 de Arabidopsis thaliana”. Instituto de Bioingeniería. Universidad Miguel Hernández. Agencia de financiación: Ayudas de la Generalitat Valenciana para grupos de investigación consolidables. Vigencia: 2015 – 2016.
PI: Víctor Manuel Quesada Pérez.
Análisis de la familia génica de los factores de transcripción mTERF en las plantas. Agencia de financiación: Generalitat Valenciana. Vigencia: 2009 – 2010.
PI: Víctor Manuel Quesada Pérez.
